Using AI to reinvent Drug Discovery: A Case Study on Recursion Pharmaceuticals [Case Studies]
Can we utilize advanced pattern-matching algorithms to bring drugs to trial more quickly
Hey, it’s Devansh 👋👋
In Case Studies, I look into specific Organizations to understand how they solve various challenges in AI/Tech. These Case Studies are meant to provide deeper insight into the different industries applying AI, to gain deeper insight into what it takes to successfully deploy AI in the wild. Shoot me a message if you come across any interesting implementations.
I put a lot of effort into creating work that is informative, useful, and independent from undue influence. If you’d like to support my writing, please consider becoming a paid subscriber to this newsletter. Doing so helps me put more effort into writing/research, reach more people, and supports my crippling chocolate milk addiction. Help me democratize the most important ideas in AI Research and Engineering to over 100K readers weekly. You can use the following for an email template.
PS- We follow a “pay what you can” model, which allows you to support within your means, and support my mission of providing high-quality technical education to everyone for less than the price of a cup of coffee. Check out this post for more details and to find a plan that works for you.
A massive thank you Marina T Alamanou, PhD for sharing her deep insights on the Tech-Bio space and to Aditi Pallod for helping me focus this article. Both your help was key in improving this article. My work would not be where it is without the constant interactions I have with all of you, and I’m excited for our future conversations.
Executive Highlights (TL;DR of the article)
Last month, I got to kick it with some people at Recursion Pharma (RXRX), a company that’s attempting to use AI to redefine the Drug Discovery process. Talking to them was super interesting, especially since tech-bio isn’t the most accessible domain to outsiders. In a nutshell, Recursion is attempting to solve a problem that is haunting the entire biotech space: Drug Discovery is becoming more expensive for lower ROI-

Drug discovery is an enormously expensive process with massive failure rates — and most of these failures happen in later-stage clinical trials when millions — or even billions — have already been invested in R&D and clinical development. Recursion is using AI to iterate every piece of the drug discovery and development workflow — adding more purpose-built data, foundational models and automation — from target identification to clinical trials.
In doing so, they’re looking to maximize the pattern-matching capabilities of AI to discover relationships between molecules and the outcomes they lead to (making them tech-bio, not biotech). If their bet pays off, Recursion will be able to mass-produce drugs for (comparative) peanuts- since they will be able to speedrun the initial screening phase that sucks up a lot of time and energy.

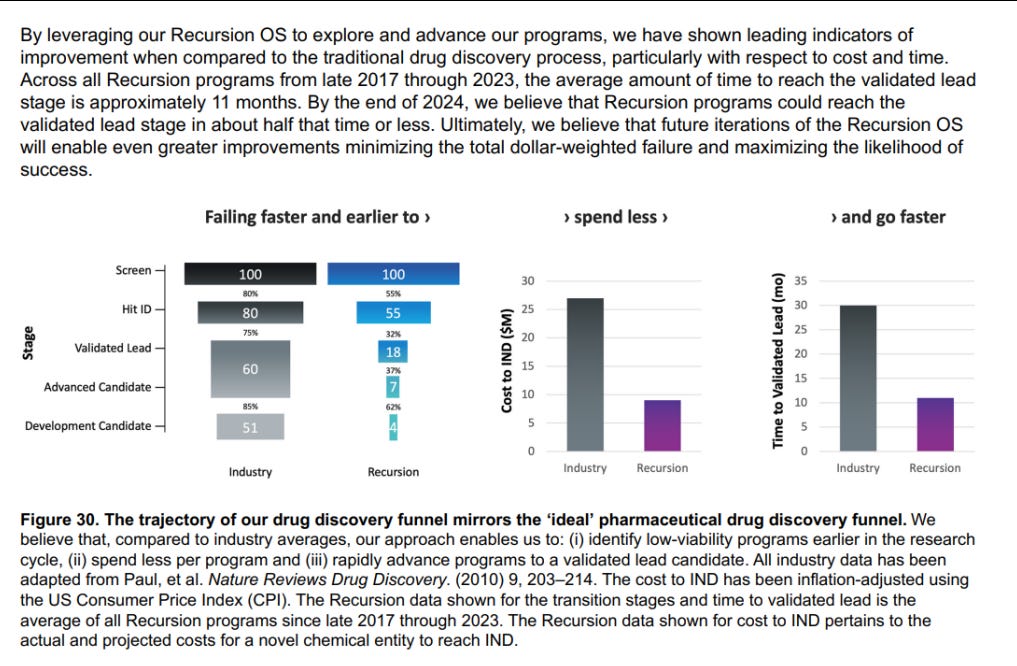
Recursion has also already demonstrated that they can massively reduce the time and cost of early-stage drug discovery — instead of years and millions of dollars to identify candidate programs and generate the data to support their predictions, they can confirm insights for tens of thousands of dollars each and in just weeks.
Other early results are promising with more program hypotheses initiated by a team of fewer people (it’s important to note that this doesn’t talk about the quality of each hypothesis, and studying the comparative ROI would be interesting)-

Using their operating system, they’ve developed a pipeline of drugs in cancer and rare diseases, in addition to over a dozen preclinical programs in the works. 7 of these AI-developed drugs in development are nearing clinical trial readouts in the next 18 months — 5 of them for Phase II trials. These readouts will ultimately determine the effectiveness of this model in delivering better drugs faster — and if (this is a pretty big if) this demonstrates ROI, then the entire drug-discovery space will experience a paradigm shift in how things are done (this might be their “Attention is All You Need” moment).
Recursion accomplishes this by running a tried and tested playbook- they execute automated experiments at very high throughput, which produces a lot of high-quality data. They process and feed this data into their AI Models, giving them the insights to pull out new possible molecules and drugs. All of this is enabled by their investments into super-computing, which allows them to yeet data at unprecedented scales.
So what does Recursion do differently? Their key innovation is in their decision to look across the entire process- from a Genome to the Final Human trials- instead of focusing their data on one specific layer like traditional setups-

To me, this is similar to the difference between diffusion and auto-regression. AR is easier (since it only looks at the next output), so it reaches high performance earlier. But if you can successfully tackle the added complexity of looking through the entire sample when generating, you get higher-quality output by using Diffusion. Recursion is betting that their super-compute-enabled Data Pipeline can pull this off and build detailed relationships across the entire pipeline instead of myopically focusing on one stage.
They are optimistic about the Log-Linear scaling shown by their models, which indicates that investing in more compute/scale will continue to raise the performance steadily for now-
I have two thoughts about this-
I’m not sure that this is the best approach. There is so much amazing research into improving Deep Learning without relying purely on scale. I wish Recursion had shared some more information on their unit economics, their evaluation of opportunity costs, estimated returns, the various protocols they have tested etc. Without having that, all I can tell you is that my recommendation to you is that you look into other alternatives before jumping into super computing.
Recursion could have tested all of this and made the judgment call that the best bet was to scale with supercomputers and allocate their cognitive resources elsewhere.
Given that I don’t have access to internal documents, I can’t tell you which it is. But I will flag that there are alternatives to this super-computing play (we discuss this a bit more at the end of the article), and any team working on this domain should be cognizant of that and not rush to copy the super-compute scaling strategy blindly.
Regardless of the reasons, there is still a lot to be gained by studying Recursion and its unique twist on this very difficult space. Below is a more comprehensive overview of the possible impacts of the Recursion Experiment-
The rest of this article will explore the following ideas in more detail:
Challenges in Traditional Drug Discovery
The pharmaceutical industry faces high failure rates, long timelines, and enormous costs. Limited understanding of complex biology, difficulty translating lab results to clinical efficacy, and a lack of open, large-scale datasets hinder progress. Traditional screening approaches often miss complex cellular interactions, especially since most analysis is done at a relatively narrow scope (going back to the conversation about focusing on one data layer vs the entire stack earlier). This focus can introduce unintended biases-
Here we performed a phenotypic CRISPR–Cas9 scan targeting 17,065 genes in primary human cells, revealing a ‘proximity bias’ in which CRISPR knockouts show unexpected similarities to unrelated genes on the same chromosome arm. This bias was found to be consistent across cell types, laboratories, Cas9 delivery methods and assay modalities…This previously uncharacterized effect has implications for functional genomic studies using CRISPR–Cas9, with applications in discovery biology, drug-target identification, cell therapies and genetic therapeutics.
As with many other fields, the industry also struggles with integrating diverse data types and interpreting high-dimensional biological data. We’ll likely see more bio-specific embeddings and techniques to handle this, which would be interesting. I have my money on Fractals, since they are key in modeling chaotic systems and our bodies would meet the criteria. Also, our body is filled with Fractals, and this is key for our health- “ Although the basis for fractal behaviour in natural systems is still unclear, its relationship to physiological state is much more pronounced. Variability has been analyzed with chaos and fractal theory in many different physiological systems, and the measure of fractality has been demonstrated to be associated with the state of health of the system”.
The RXRX crew is banking on the following to tackle these problems-
Recursion’s Data Generation and AI Analysis:
Recursion generates massive cellular imaging datasets using high-throughput microscopy and genetic/chemical perturbations (their lab looks super cool from the outside). This is stacked with deep learning and self-supervision, extracting complex features from millions of images to enable a more comprehensive understanding of cellular biology.
This is shown beautifully in their AI’s ability to distinguish between very subtle differences between cells-

And in the research below-
Our results show that both CNN- and ViT-based masked autoencoders significantly outperform weakly supervised baselines. At the high-end of our scale, a ViT-L/8 trained on over 3.5-billion unique crops sampled from 93-million microscopy images achieves relative improvements as high as 28% over our best weakly supervised baseline at inferring known biological relationships curated from public databases
-Masked Autoencoders are Scalable Learners of Cellular Morphology
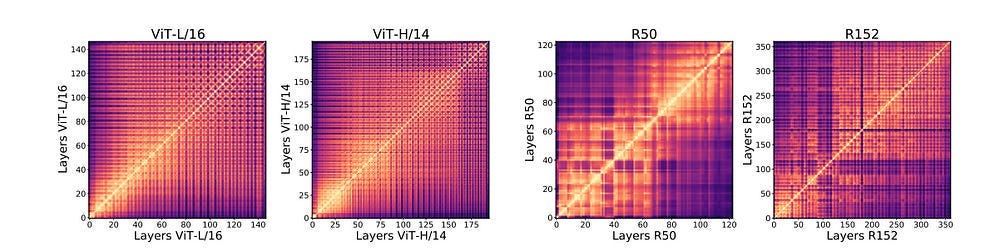
The superior performance of ViTs over CNNs is interesting and might be attributed to two factors. Firstly, it could just be the quality of the features pulled out Vision Transformers, which objectively see differently- “Representation structure of ViTs and convolutional networks show significant differences, with ViTs having highly similar representations throughout the model, while the ResNet models show much lower similarity between lower and higher layers. We plot CKA similarities between all pairs of layers across different model architectures. The results are shown as a heatmap, with the x and y axes indexing the layers from input to output. We observe that ViTs have relatively uniform layer similarity structure, with a clear grid-like pattern and large similarity between lower and higher layers. By contrast, the ResNet models show clear stages in similarity structure, with smaller similarity scores between lower and higher layers.”
However, this isn’t the only possible reason. The slept-on “A ConvNet for the 2020s”, posits that a large part of the reason that Transformers have been outperforming CNNs in Vision-related tasks has been the superior training protocols used by Transformers (which are a newer architecture). Thus, by improving the pipeline around the models, they argue that we can close the performance gap between Transformers and CNNs. They’re able to close the performance gap by applying several modernizing tricks-
Constructed entirely from standard ConvNet modules, ConvNeXts compete favorably with Transformers in terms of accuracy and scalability, achieving 87.8% ImageNet top-1 accuracy and outperforming Swin Transformers on COCO detection and ADE20K segmentation, while maintaining the simplicity and efficiency of standard ConvNets.
I’d love to see how much each of the above factors play a role. Given the nature of this problem, I’d also be interested in how Contrastive-style techniques like Triplet Loss would impact the performance of this system (my guess is that they would do very well)-

Then, we have MolE, a molecular foundation model that adapts the DeBERTa architecture to work with molecular graphs. It employs a novel two-step pretraining strategy: “The first step of pretraining is a self-supervised approach focused on learning chemical structures, and the second step is a massive multi-task approach to learn biological information. We show that fine-tuning pretrained MolE achieves state-of-the-art results on 9 of the 22 ADMET tasks included in the Therapeutic Data Commons”-

All of these combine to give Recursion the ability to hit clinical trials much quicker than their competitors (18 months to the industry standard of 42)-
FYI- Recursion has and is going to invest heavily in open-sourcing their data and models. Phenom-Beta and the RXRX3 database are publicly available. More info on these here: https://www.drugdiscoverytrends.com/recursion-phenom-beta-phenomics/. This is a great opportunity for anyone who wants to get involved, and I’d strongly suggest reaching out to Recursion to check out their Open Source initiatives. You can also find LinkedIn over here and/or their Twitter/X here.
To succeed, RXRX will have to solve several challenges. On a technical front, the usual suspects in model interpretability, biological variability, and translating results to clinical success have Yamcha’d many promising ideas, and RXRX will have to deal with them. Given their work’s sensitive (and regulated) nature, combined with the current regulatory environment, I would imagine AI transparency to be a particularly important facet. We’ll discuss this more in the challenges section, but Medical AI, in particular, needs to put a lot of emphasis on the transparency/AI explainability side of the process.
As an interesting philosophical tangent, it is interesting to see how much money/attention is paid to fixing disease as opposed to augmenting humans. Given their focus on genotype-phenotype mapping, Recursion could have decided to follow the wisdom of the dinosaur below-

AFAIK lots of people have dedicated lots of money and time (even making it their life mission) to boring things like solving diseases and saving lives. No one has put anything into giving us laser eyes and finger canons. This asymmetry in resource allocation is interesting, and I’d be curious to hear what you think it says about us as a species.
On that note, if any investors want to give me their money to fix this imbalance, I promise it all on trying to develop cool things like the Stand-Giving arrow from Jojo (or the ideas mentioned above). Cancer be damned. Shoot me a message (links below) if this vision interests you.
And that, my special ones, is a very high-level overview of the Drug Discovery space and how Recursion Pharma is trying to change it. We will now get into the details, starting first by understanding the challenges in the Drug Discovery Domain.
PS- My family was very worried about me failing biology and chemistry in high school, so if you think I’ve misunderstood or missed anything important, please feel free to let me know.
I provide various consulting and advisory services. If you‘d like to explore how we can work together, reach out to me through any of my socials over here or reply to this email.
I put a lot of work into writing this newsletter. To do so, I rely on you for support. If a few more people choose to become paid subscribers, the Chocolate Milk Cult can continue to provide high-quality and accessible education and opportunities to anyone who needs it. If you think this mission is worth contributing to, please consider a premium subscription. You can do so for less than the cost of a Netflix Subscription.
Why Deepmind hasn’t solved Drug Discovery Yet
Section Summary- Traditional drug discovery faces high failure rates, long timelines, and enormous costs. Key technical challenges include limited understanding of complex biology (which leads to a reliance on oversimplification to solve problems), difficulty translating lab results to clinical efficacy, and a lack of open, large-scale datasets. These issues necessitate novel approaches to revolutionize the drug discovery process.
If you’re like me, you’ve probably looked at all the hype about developments like AlphaFold and wondered why they haven’t led to the revolution that you’d expect. Turns out there are a few reasons for this.
Obviously, you’re dealing with very very high costs- “Estimates of total average capitalized pre-launch R&D costs varied widely, ranging from $161 million to $4.54 billion (2019 US$)… Our analysis identified a trend of increasing R&D costs per NME over time… We found no evidence of an increase in suitability scores over time.”
At the root of these high costs is our limited understanding of complex biological systems. The human body’s intricate network of interconnected processes means diseases often arise from multifaceted interactions rather than single-point failures. Traditional approaches, focusing on isolated protein targets or simplified cellular assays, struggle to capture this complexity.
We add to this oversimplification by using proxies to assess the impact of drugs. Proxies in drug discovery have two drawbacks-
Obviously, since we can’t jam every experimental drug into people to study their impacts, proxies in Drug Discovery are particularly distant from our desired measurement (it is difficult to forecast how drugs would interact with each other/other lifestyle decisions). I wonder if worsening economic conditions and mental health will lead to looser regulations on human testing.
Biological systems can have delayed feedback, which is extremely hard to account for in experiments. How do you test and account for unintended long-term changes from a particular molecule?

Our oversimplification doesn’t end here. Traditional screening approaches also face limitations in capturing the full spectrum of cellular responses to potential drugs. High-throughput methods often rely on simplified readouts, potentially missing subtle, system-wide effects that ultimately determine a compound’s therapeutic potential.

This leads to a distributional shift between lab results and IRL drugs since the data we work on doesn’t fully represent the system. Hence, many candidates who show promise in controlled laboratory conditions fail at later stages of testing.
Our data troubles don’t end here. As a field, tech-bio needs more open, large-scale biological datasets. The proprietary nature of pharmaceutical research often leads to data silos, with valuable information — especially negative results — remaining hidden from the broader scientific community. This lack of data sharing hampers progress and leads to duplication of efforts.

Even those with access to large scales of data have their burdens to bear. The challenge of integrating and interpreting the ever-growing volume of biological data from various -omics fields presents another significant obstacle. Researchers are often faced with a deluge of information but lack the tools to effectively mine it for meaningful patterns and relationships.
Lastly, the slow iteration cycles of traditional wet lab experiments create a bottleneck in the drug discovery process. Each round of testing requires significant time and resources, leading to worse performance.
This is what Recursion is trying to tackle. Let’s talk about how.
Using AI and Big Data for Drug Discovery
Section Summary: Recursion tackles traditional drug discovery challenges through massive-scale cellular imaging, advanced AI analysis, and integration of multiple data types. Their platform generates rich phenotypic data, uses deep learning to extract complex features, and leverages this information for biological insights and drug target identification.
The crux of Recursion’s approach can be summarized as follows-
Generate a lot of high-quality data from across the data layers.
Run this through a supercomputer to do cross-sectional pattern matching.
Roll in money. GG EZ.
The company’s data generation pipeline centers on high-throughput cellular imaging, specifically utilizing the Cell Painting assay. This method allows for the visualization and quantification of multiple cellular components simultaneously, providing a rich, multidimensional view of cellular state. Recursion has scaled this approach to an unprecedented level, generating millions of cellular images across various cell types and conditions.
For comparison, this is how Recursion’s Biological Dataset compares to some other heavy-hitters in the Computer Vision Space-
As mentioned in the tl;dr, Recursion runs this dataset through tuned Vision Models to teach them the relevant structures of Cells. Their ability to reconstruct from partial samples is pretty impressive-
For those of you that prefer tables, you can catch their performance below-

The company’s approach doesn’t stop at image analysis. Recursion integrates multiple data types, including transcriptomics and chemical structures, to build a more comprehensive understanding of biology and drug-target interactions. This multimodal integration allows for more robust predictions and insights.
By analyzing similarities between cellular perturbations across their massive datasets, Recursion’s platform can infer biological relationships, recapitulate known pathways, and discover novel connections.
Their approach has enabled Recursion to find important flaws in contemporary methods. Take a look at the figure, which presents a genome-wide heat map of phenotypic similarities between gene knockouts. The striking diagonal patterns reveal that knockouts of genes located close together on the same chromosome arm tend to produce more similar cellular phenotypes, an effect termed “proximity bias”. This unexpected similarity between nearby genes is consistently observed across different datasets (rxrx3 above the diagonal, cpg0016 below), highlighting a previously uncharacterized confounding factor in CRISPR-Cas9 screens that spans cell types and experimental setups.
Once we identify this bias, we can correct it using a geometric method called “chromosome-arm correction”. Here’s a brief summary of their approach:
1. For each chromosome arm, they calculate the average feature vector across all unexpressed genes on that arm.
2. This average vector is then subtracted from the feature vectors of all genes located on that arm.
3. Unexpressed genes are defined based on RNA sequencing data, specifically those with very low expression levels (zFPKM < -3.0) before any CRISPR-Cas9 treatment.
This method effectively removes the shared background signal within each chromosome arm while preserving the unique biological signals of individual genes. By using unexpressed genes as a reference, they aim to capture and subtract out the non-specific effects related to chromosomal location without interfering with genuine biological relationships.
This correction significantly reduces the proximity bias effect both globally and for each chromosome arm, while maintaining or even improving the ability to detect known biological relationships across the genome.
Additionally, the potential false positive driver genes for specific cancer subtypes discussed in the previous section are reduced with six of the nine highlighted genes no longer showing a subtype-specific dependence
-This is a pretty cool outcome
The image below, shows the impact of this correction, with the similarities in the bottom triangles being significantly lower than the top half-

So far, these results have been very promising but not truly game-changing. However, they are good enough to convince Recursion to double down on their bet on using Super-Computing. Let’s talk about their newest Super-Computer and Partnership with Nvidia that will help them push the scale even further.
BioHive 2 and Nvidia’s 50 Million USD Investment
As mentioned earlier, Recursion’s team is betting big on Log-Linear Scaling demonstrated by their experiments to push their boundaries. By partnering with Nvidia, they’ve built a new super-computer- BioHive-2 (the fastest super computer in Pharma)- which allows them to speed up their Data Analysis significantly. They’re hoping this added computational firepower will help turn their Lamine Yamal into Messi.
I’m torn on this. From what I’ve read in their work, I don’t think the RXRX team has fully maxed out the potential of embeddings and active learning, so I think investing this heavily into a new super-computer might be premature.
For example, a drop in error from 3% to 2% might require an order of magnitude more data, compute, or energy. In language modeling with large transformers, a drop in cross entropy loss from about 3.4 to 2.8 nats requires 10 times more training data . Also, for large vision transformers, an additional 2 billion pre-training data points (starting from 1 billion) leads to an accuracy gain on ImageNet of a few percentage points…we could go from 3% to 2% error by only adding a few carefully chosen training examples, rather than collecting 10x more random ones
-Meta’s “Beyond neural scaling laws: beating power law scaling via data pruning” is one of many great publications that’s looking at beating power-law scaling.
The skeptic in me would call this an attempt to cash in on the Nvidia/Deep Learning hype. We’ve seen that story before, such UnderArmor’s failed attempt to cash in on the BigData/Digitization hype by acquiring a bunch of apps w/o any regard for business fundamentals
However, it is possible that they tried everything, failed, and never bothered publishing the results. Or they’ve decided that the attention of their AI teams would be better focused on other avenues.
Or maybe I’m just a super-computer-less peasant who should STFU and let the bros cook.
Whatever the case, the results seem promising enough to pay attention-
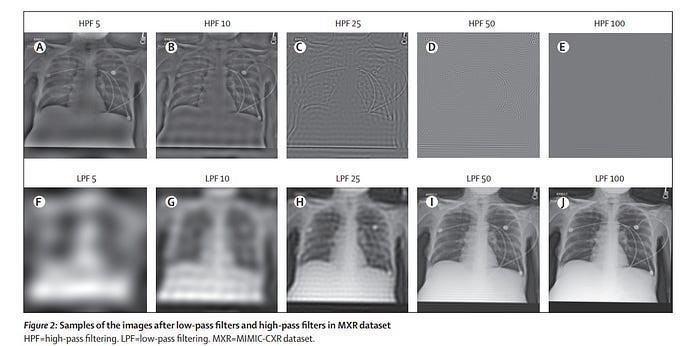
As Recursion continues to scale, I think they’ll likely need to invest more into (and highlight) their endeavors in AI Transparency. Especially given their reliance on Deep Learning, which while promising, has too much we don’t understand. To take an example close to home (AI Medical Imaging), let’s take the paper, “AI Recognition of Patient Race in Medical Imaging: A Modelling Study”. The authors of the paper demonstrate something very interesting (and worrying)- we can train simple deep-learning classifiers to infer the race of people by looking at pictures of their lungs (a task considered impossible by medical experts). Most surprisingly, the classifiers are remarkably robust to noise, maintaining an ~80% classification accuracy even when passed through noise that turns the image into what looks like random noise to medical experts (look at HPF 100)-
Because this point is easily misunderstood, I want to clarify something: It’s not a problem that AI has this amazing classification ability. What worries me is that experts don’t know why and that there might be tons of other hidden correlations that AI might pick up on w/o us knowing. To successfully deploy safe AI in such sensitive and heavily regulated fields- it’s important to start investigations bringing such correlations to the light. This is why risk-averse industries like Finance have been hesitant to adopt to Deep Learning-
Disclaimer: I’m a SWE and an ML enthusiasts, but not an expert by any means. Seems promising more so in terms of explainability than actual performance. I currently work in a heavily regulated industry and explainability is the bottleneck for adoption of ML models.
- Cole Sawyer, Capital One, when talking about why he was excited about Kolmogorov–Arnold Networks
The only way to address this is to conduct large-scale experiments to uncover these relationships. If nothing else, the team at Recursion seems to have the right foundations to do so.
And that is my overall assessment of Recursion. Despite my critiques, I think they’re worth paying attention to (hence this article). They have a very good team, some exciting partnerships, and a clear enough vision to push on their “go big or home homeless bet.” Regardless of whether they fully succeed in what they’re trying to accomplish- we will learn a lot by following the Recursion Experiment.
If you have any thoughts on Recursion or their approach with Tech-Bio, I’d love to hear them.
If you liked this article and wish to share it, please refer to the following guidelines.
That is it for this piece. I appreciate your time. As always, if you’re interested in working with me or checking out my other work, my links will be at the end of this email/post. And if you found value in this write-up, I would appreciate you sharing it with more people. It is word-of-mouth referrals like yours that help me grow.
Reach out to me
Use the links below to check out my other content, learn more about tutoring, reach out to me about projects, or just to say hi.
Small Snippets about Tech, AI and Machine Learning over here
AI Newsletter- https://artificialintelligencemadesimple.substack.com/
My grandma’s favorite Tech Newsletter- https://codinginterviewsmadesimple.substack.com/
Check out my other articles on Medium. : https://rb.gy/zn1aiu
My YouTube: https://rb.gy/88iwdd
Reach out to me on LinkedIn. Let’s connect: https://rb.gy/m5ok2y
My Instagram: https://rb.gy/gmvuy9
My Twitter: https://twitter.com/Machine01776819